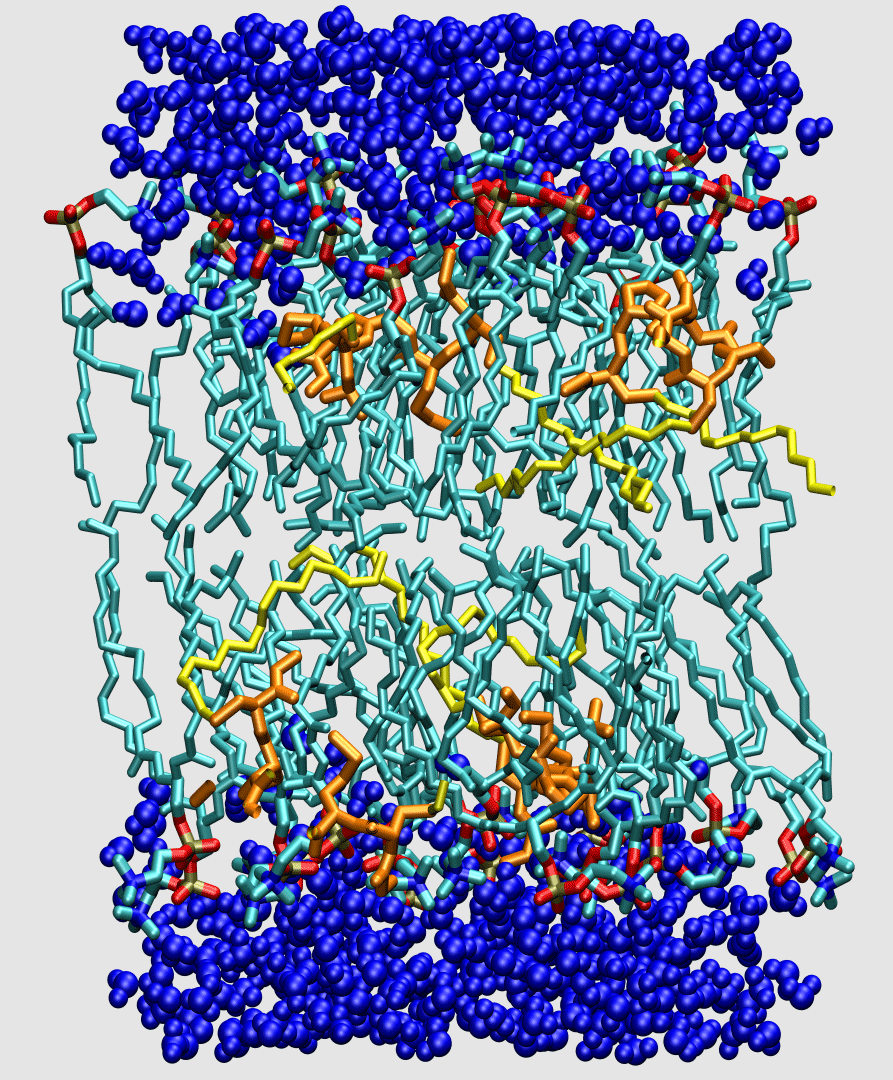
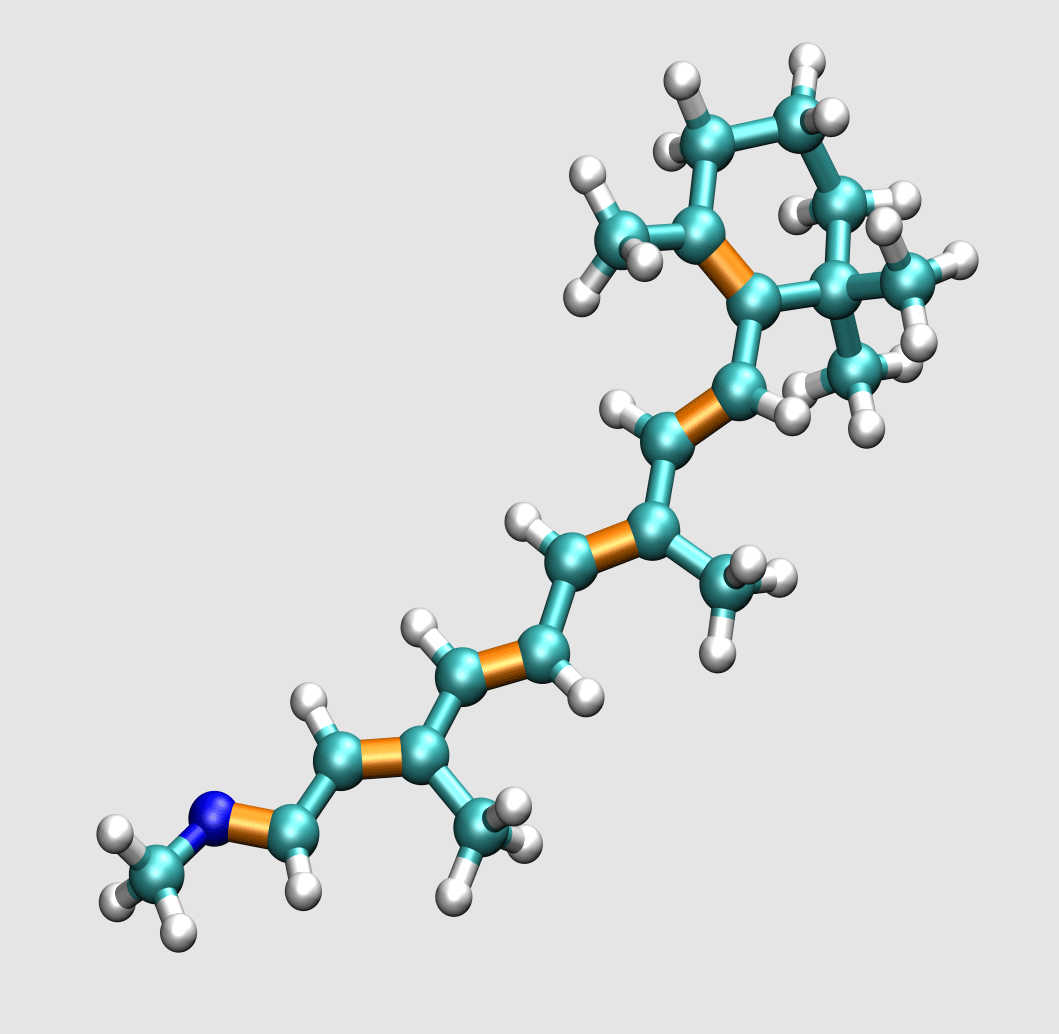
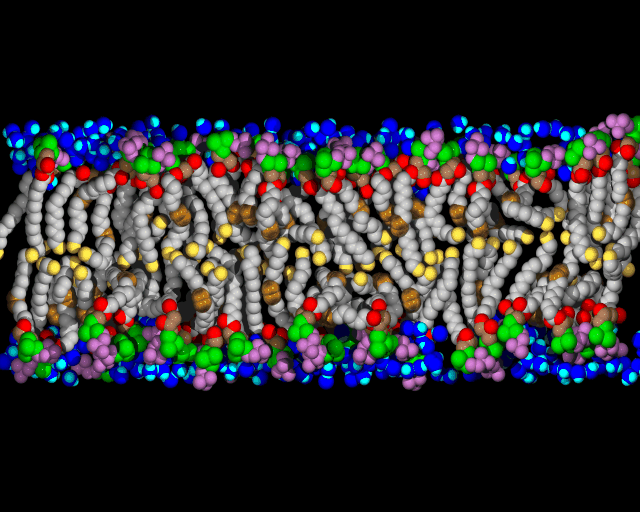
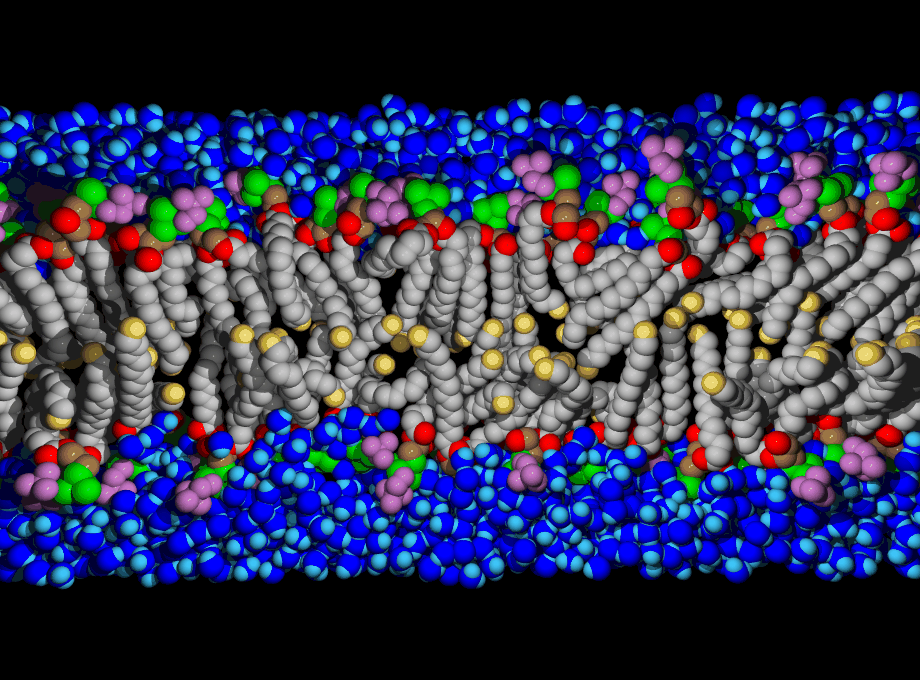
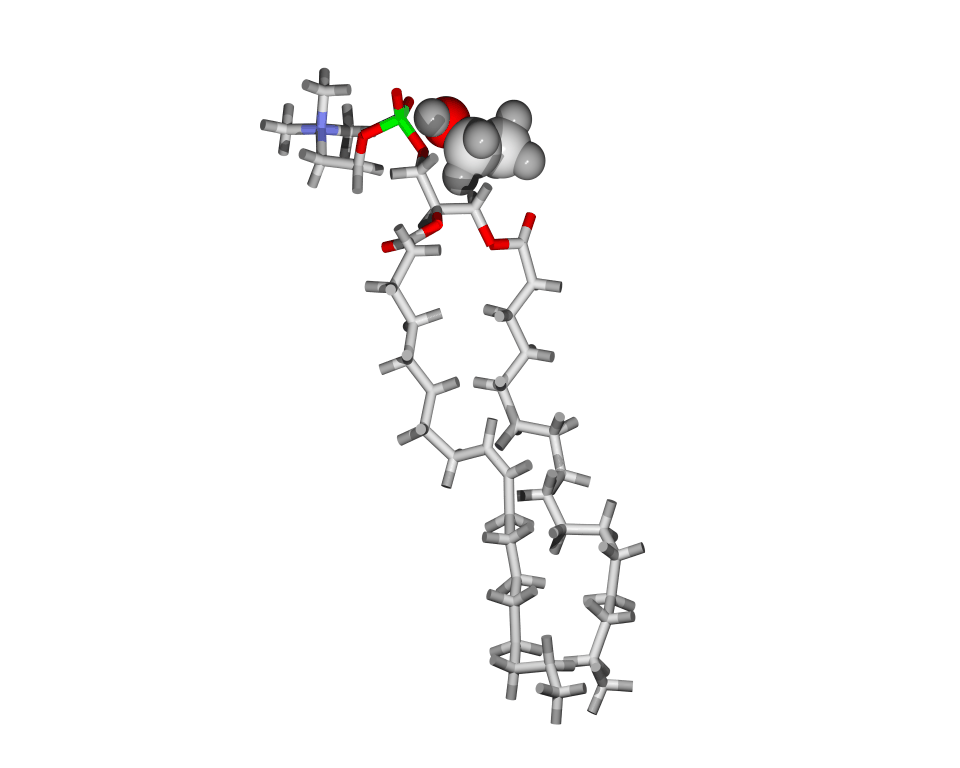
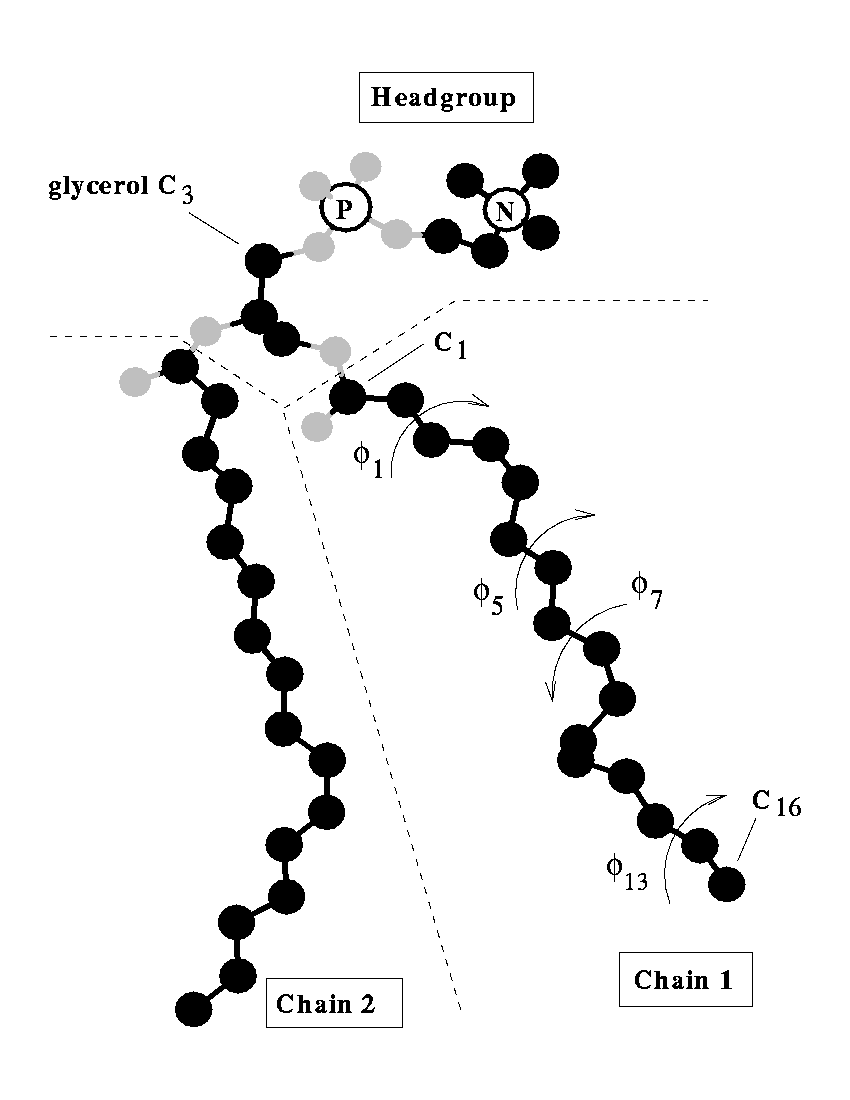
Lipids are the building blocks for biological membranes. They are characterized based on three basic components: (1) a dipole headgroup that is hydrophilic
or "water loving", (2,3) and two hydrocarbon chains that are hydrophobic
or "water hating". These chemical properties give the lipid bilayer a preferred construct: the headgroups stay near the water and the chains hide in the interior of the membrane.
Most lipids investigated by the lab are phospholipids characterized by the presence of phosphate (P) and choline (N) subgroups in the headgroup. The tails are classified by the number of carbons, and position and number
of carbon-carbon double bonds which increase flexibility. A glycerol backbone links the headgroup and hydrocarbon tails.

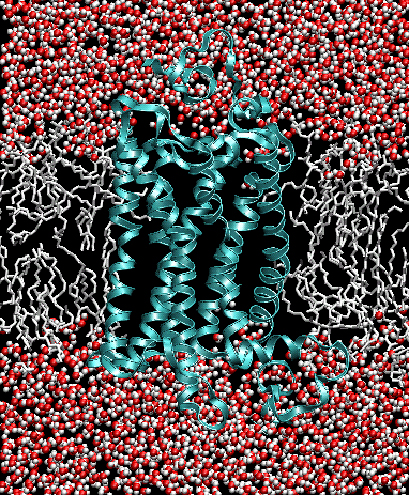
Biological membranes are composed of two layers of lipids organized into sheets. They form a nearly impermeable barrier to water-soluble molecules, especially ions. Membrane-bound proteins
interact with lipids to establish a controlled mechanism for signaling across the barrier and for diffusion
of material through the barrier. Bilayers can also act as an anchoring point for proteins.